
New area of research in our lab: Peptide Antibiotics development
A new area of research in our lab, is the development of novel peptide-based antibiotic compounds.
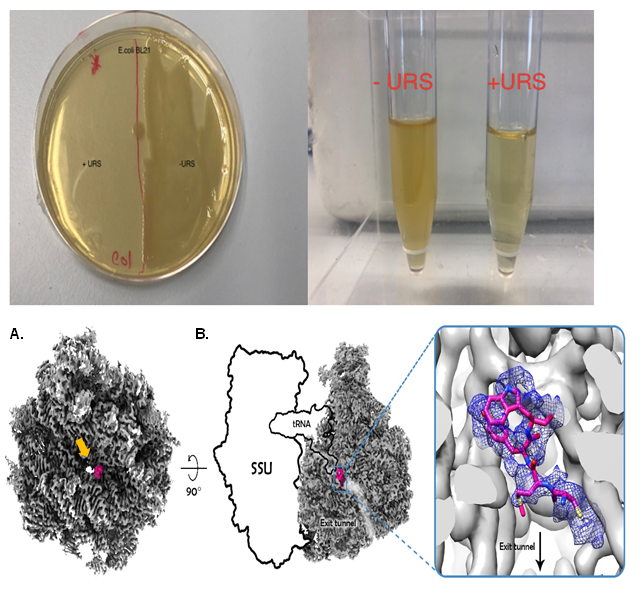
Using a novel bioinformatic method, we discovered amino acid sequences that are underrepresented in the bacterial proteome – we call them URS sequences. that when synthesized as short peptides inhibit protein translation by ribosomal arrest.
The URS’s effect on the cell viability was examined in vivo, using expressed proteins with embedded URS sequences, and we observed that when the URS-protein was expressed, ribosomal activity was significantly reduced, leading to protein synthesis inhibition, followed by cell damage or death. The study determined that the URS inhibitory action results from interfering with the ribosome exit tunnel, leading to ribosomal stalling[1].
When isolated synthetic peptides with the URS sequences were added to growing bacterial culture we were able to show inhibitory action both in vivo (bactericidal activity) and in vitro (inhibition of translation.
Our aim is to determine the inhibitory mechanism to maximize the URS peptides activity, then we will move to the first trials of URS peptides as an antibiotic for pathogenic bacteria.
We are also working on the atomic structure determination of ribosome -URS complex (stalled ribosome), by either X-Ray crystallography or cryo-Electron Microscopy.
Structure determination will allow us to understand more about the molecular interaction between the URS and the ribosome.
1. Navon, S. P., Kornberg, G., Chen, J., Schwartzman, T., Tsai, A., Puglisi, E. V., … & Adir, N. (2016). Amino acid sequence repertoire of the bacterial proteome and the occurrence of untranslatable sequences. Proceedings of the National Academy of Sciences, 113(26), 7166-7170.
Read MSc Thesis
Tareq's Linkedin Profile

Prof. Noam Adir and Tareq looking at a protein structure

Tareq – Prize Winner at The Faculty Research Day 2022

Tareq with a Petri Dish

