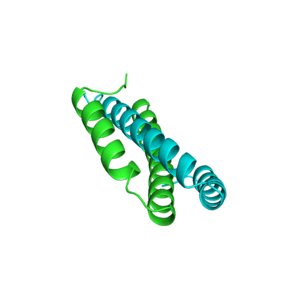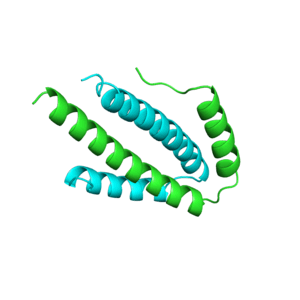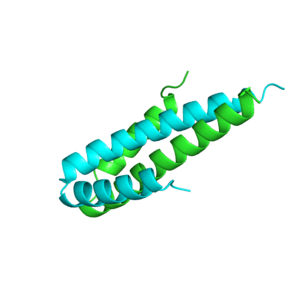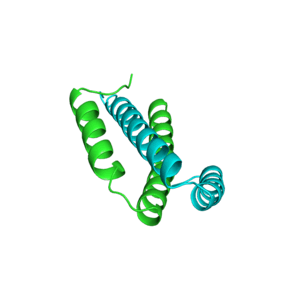
Structures
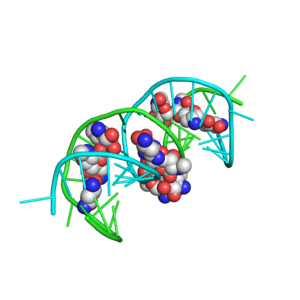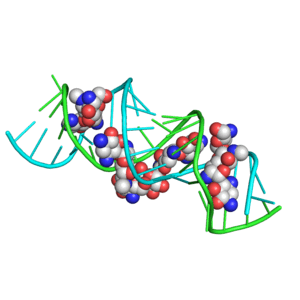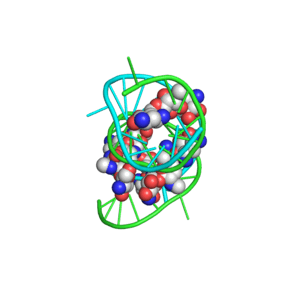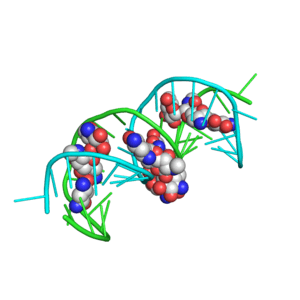
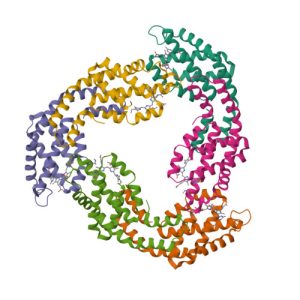
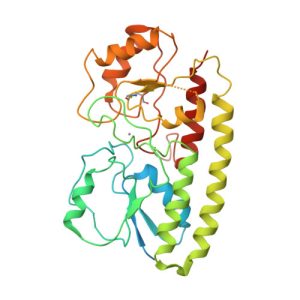
1R85
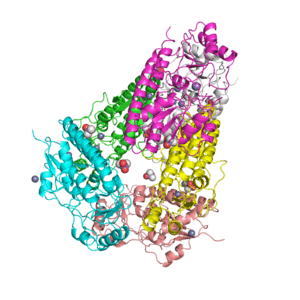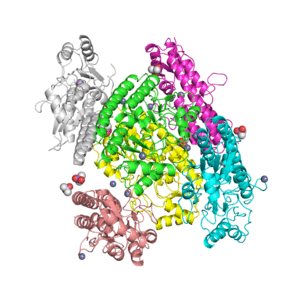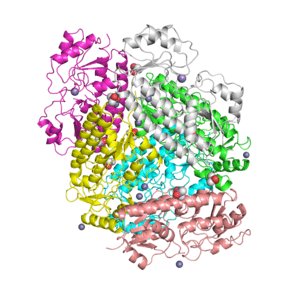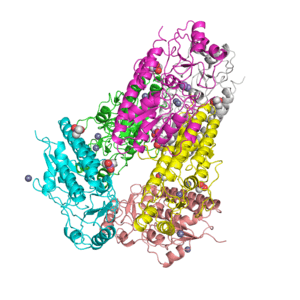
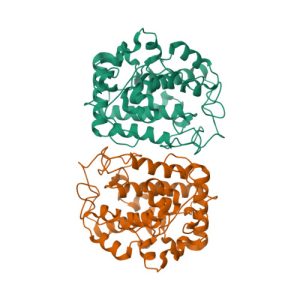
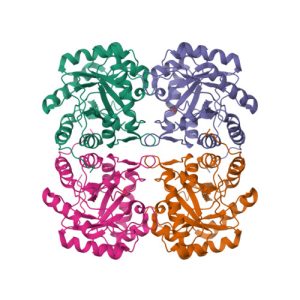
Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6): The WT enzyme (monoclinic form) at 1.45A resolution
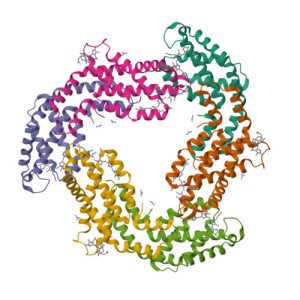
1R87
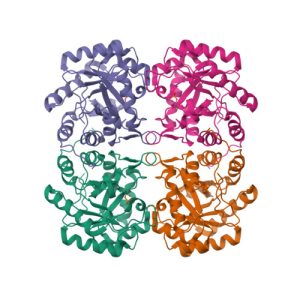
Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6, monoclinic form): The complex of the WT enzyme with xylopentaose at 1.67A resolution
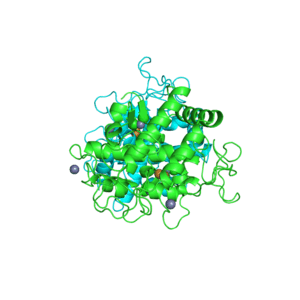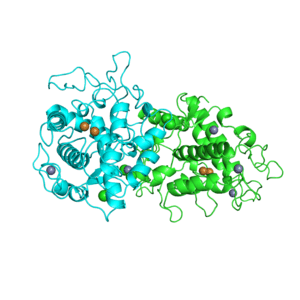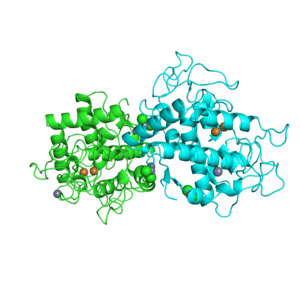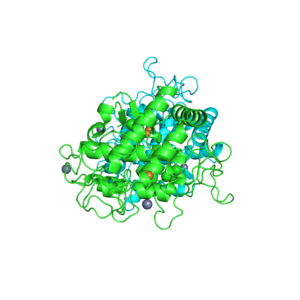
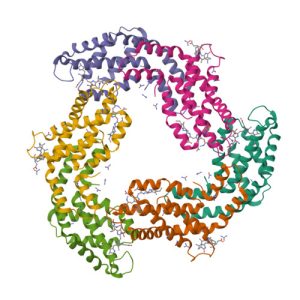
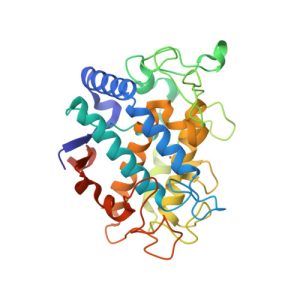
1PHW
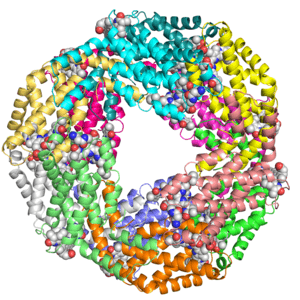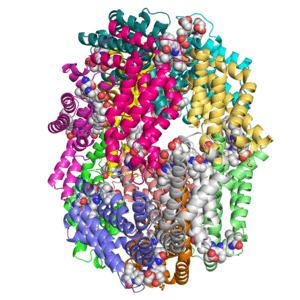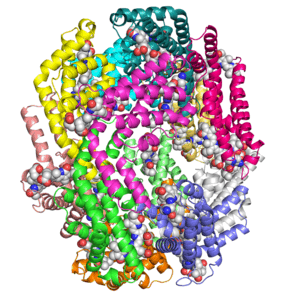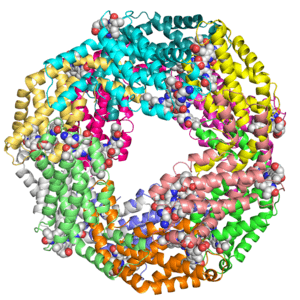
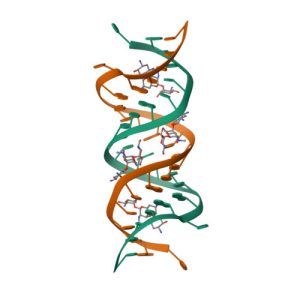
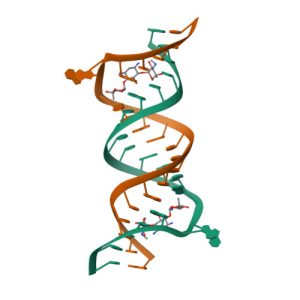
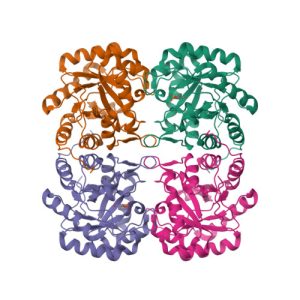
Crystal structure of KDO8P synthase in its binary complex with substrate analog 1-deoxy-A5P
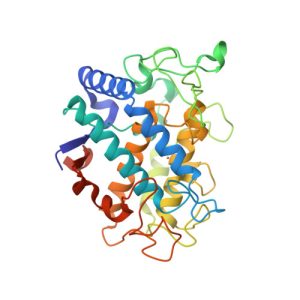
1G7U
Crystal structures of KDO8P synthase in its binary complex with substrate phosphoenol pyruvare
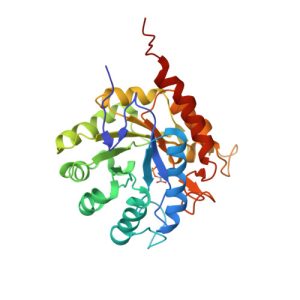
1G7V
Crystal structures of KDO8P synthase in its binary complex with the mechanism-based inhibitor
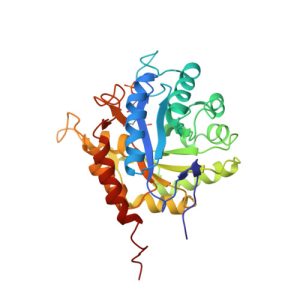
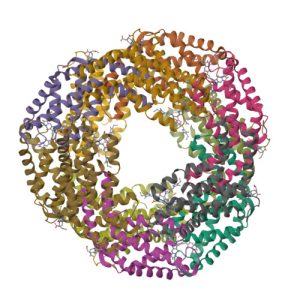
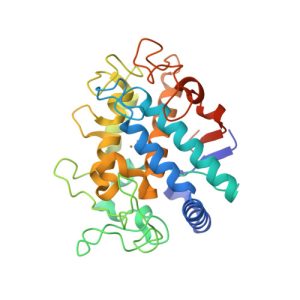
1I7Y
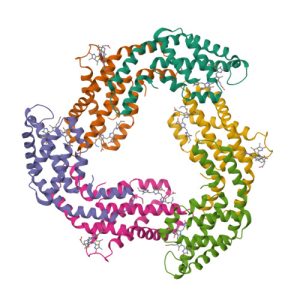
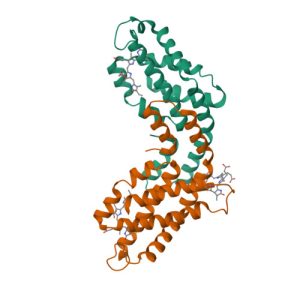
Structure of c-phycocyanin from the thermophilic cyanobacterium Synechococcus vulcanus at 2.5 A: structural implications for thermal stability in phycobilisome assembly