
Designing a Phycobiliprotein Based Exciton Energy Transfer System
The T. vulcanus Phycocyanine (PC) is one of the proteins that form the rod structure of the T. vulcanus PBS. Previous research showed that isolated and purified PC trimers can undergo a self-assembly process and form a bundle of nanowires.
Those bundles retain their ability to transfer energy along the wire.
While this fact on its own can have interesting consequences on our understanding of how the PBS forms naturally in the cell, we intend to use those bundles to investigate the Excitonic Energy Transfer (EET) capabilities of PBPs.
By considering the semiclassical interpretation of EET using the Förster resonance energy transfer (FRET) model, we can imagine the energy transfer as the excited state passing through a line of PC “beads” until it reaches its target.
While this is a serviceable description of EET, other studies showed that the EET in the photosynthetic system exhibits quantum characteristics that are ill-fitting to the FRET model.
To investigate the quantum characteristics of the photosynthetic EET, we must design a new system based on the earlier PC bundle system. The main challenge that the new system will tackle is the move from bundles of PC nanowires to a single PC nanowire that will form our one-dimensional EET system. Since disorder in a system can disrupt quantum effects, we expect that a simpler system can better exhibit nontrivial quantum effects, such as delocalization of the excited state along multiple units in the PC chain.
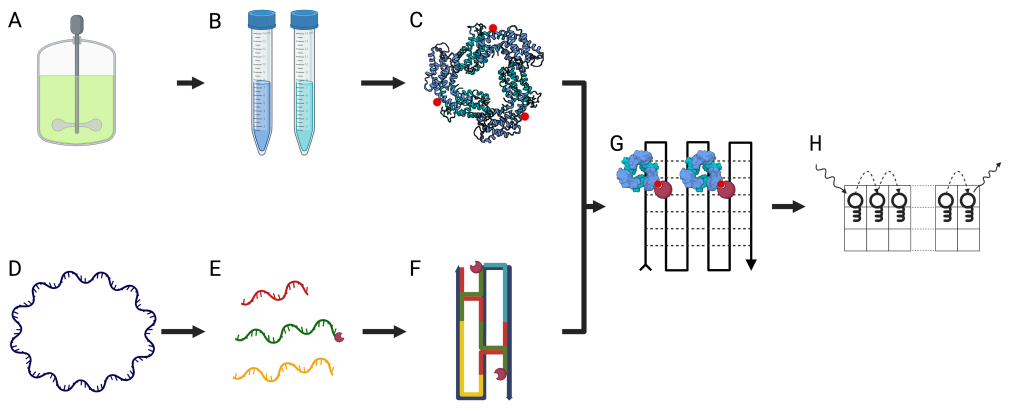
Figure 1 – A scheme showing the different steps for the preparation and assembly of the Quantum Nano-Grid system (QNG). (A) T. vulcanus cells are continuously grown in a dedicated growth container, harvested by centrifugation, and frozen for later use. (B) Phycobiliproteins are extracted and purified from the T. vulcanus cell. (C) The phycobiliproteins are biotinylated using the exposed cysteine residues. (D) (E) A large ssDNA template is chosen, and short ssDNA staples are designed according to the desired attributes of the final grid. Specific strands are attached to streptavidin to facilitate the formation of activated squares in the finalized grid. (F) The ssDNA staples fold the template according to the base pairing of complementary regions in the template. The streptavidin position is dictated by the area complementary to the staple to which they were attached. (G) The biotinylated phycobiliproteins are bound by the strong association between the biotin and streptavidin to their target position in the QNG. (H) The EET rates and distance can be calculated by exciting a specific point on the QNG and measuring the location and spectral properties of the emission. Using this modular approach to EET measurement, we can explore the quantum properties and interactions of different phycobiliproteins.
Guy's Linkedin Profile


